Development of Computational Algorithms
The efficient sampling of the conformational space of molecules or the study of their kinetics require sophisticated algorithms. Combining the concepts from statistical mechanics with proper programming we create the tools which can help to study complex systems.
Genetic networks
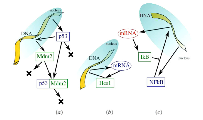
DNA and proteins build a complicated regulatory network in the cell. We study the physical features of such network and the dynamics of protein concentrations, emphasising the role of delay between the different nodes of the network.
Molecular evolution
The evolution of amino acid sequences during millions of years is the mechanism which produces the proteins of present organisms. The resulting sequences build out the most robust and abundant set of experimental data about proteins. We use again simplified models to study protein evolution, from the point of view of dynamics in the space of sequences.
Protein aggregation
Simplified models are also used to study the interaction of different proteins. Sometimes this interaction gives rise to highly ordered structures called fibrils, as in the case of insuline or of the the beta-amyloid, a protein involved in the development of Alzheimer’s disease. We study the formation of such aggregates in order to understand their origin and to design molecules to prevent the aggregation.
Thermodynamics and dynamics of protein folding
Proteins are complex systems, with many degrees of freedom, little symmetries and complicated interactions. Nonetheless, they display a unique (i.e., zero entropy) equilibrium state in a large range of temperatures and are able to reach it in a short time (typically, from microseconds to seconds). By means of simplified models, from minimal lattice models to more realistic descriptions of the protein chain, we investigate the mechanism which confer to proteins these peculiar features.
Structure of chromatin
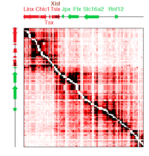
The arrangement of chromatin in the cellular nucleus is not random. Making use of polymer physics and statistical mechanics, we try to to interpret experimental data, obtained from chromosome-capture and fluorescence experiments, to understand the three-dimensional organization of chromatin and its relationship with transcriptional activity.