Our research
The Sattin Group applies organic chemistry to chemical biology problems and molecular recognition events.
When probed and analysed in enough detail molecular recognition events allow a deeper understanding of many biological processes in both physiological and pathological conditions.
Our group is focused on the design and synthesis of small molecules tailored to interact with specific protein targets. In particular, we are currently working on the inhibition of a survival bacterial enzyme to hamper bacterial persistence, the allosteric regulation of the chaperone protein Hsp90 and the modulation of self and non-self recognition via protein-carbohydrate interactions.
Probing the molecular mechanisms of bacterial persistence


Chronic and recurrent infections paired with antimicrobial resistance are rapidly becoming a serious issue in our worldwide-interconnected society. The need for new approaches and effective antimicrobials is pressing.
Bacterial persisters are an elusive phenotype that play a starring role in chronic infections, even though they are not genetically resistant to antibiotic treatment. They simply appear to shut down their metabolism becoming dormant and insensitive to current drugs. This bacterial reservoir is able to sustain pathogen survival and resurgence.
We are focusing our attention to the role played by (p)ppGpp intracellular accumulation as one of the working hypotheses for persisters formation. In particular, we are designing and synthesizing small molecules tailored to gain control over RSHs (RelA/SpoT Holomogues), a superfamily of enzymes that regulate (p)ppGpp homeostasis.[1, 2]
These small molecular tools will be used to test the hypothesis and to challenge complementary survival mechanisms.
Figure 1: Rendering of bacterial cells.
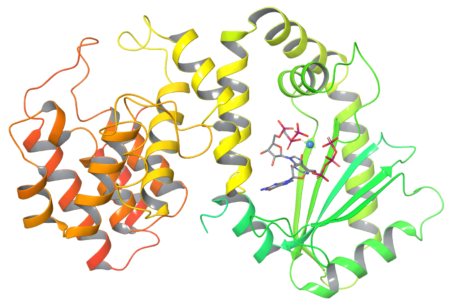
Figure 2: 3D Structure of the bacterial stress response protein RelSeq (Streptococcus equisimilis) in its modelled pre-catalytic state, bound to GTP, ATP and Mg2+.
Carbohydrates and Glycomimetics
Protein-carbohydrate interactions are the triggering step of a large variety of biochemical processes both in physiological and pathological conditions. Of special relevance is the ability of our immune system to efficiently discriminate between self and nonself through protein-carbohydrate interaction with exquisite specificity, and trigger the appropriate immune responses. Nonetheless, some pathogens, during the adhesion step, can escape this checkpoint by exploiting specific molecular receptors (e.g. DC-SIGN). On the other hand, malfunctioning of the sentinel receptors in charge of dampening the body’s reaction to self antigens (e.g. Siglecs) can cause an aberrant response at the basis of several autoimmune diseases.
We are involved on this front in different collaborative projects. We design and synthesize mono- and multivalent glycomimetic structures able to interfere with these molecular recognition events.
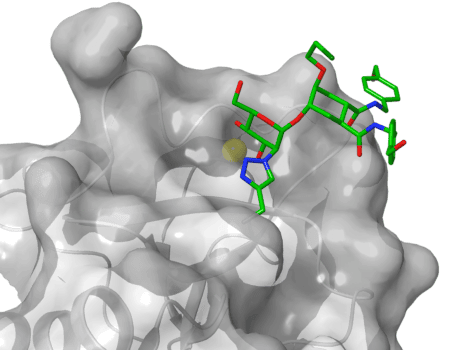
Figure 1: X-ray crystal structure of the C-type lectin DC-SIGN in complex with one of the designed glycomimetics (6ghv.pdb).
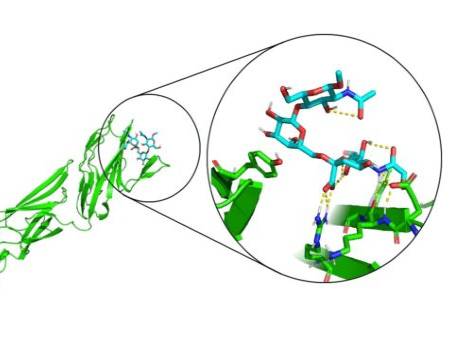
Figure 2: Human Siglec-2 (CD-22) bound to its natural trisaccharide ligand Neu5Ac-α-(2,6)-Gal-β-(1,4)-GlcNAc.
RNA-based therapeutics
RNA-based therapeutics comprise a rapidly expanding category of drugs, with the potential to access personalized medicine and traditionally “undruggable” targets in a cost-effective manner. The development of such drugs, both antisense (RNAi) and message (mRNA) RNAs, requires their spatiotemporal tracking for, e.g., target validation, residence time, off-target effects, translation, and degradation.
In this framework we are developing fluorogenic probes to perform either passive tracking of the RNA-drug, or its optical detection in response to specific steps of its uptake and processing.
This research is funded through:
PRIN 2022 PNRR

National center for gene therapy and drugs based on RNA technology

Spoke9: From target to therapy: pharmacology, safety and regulatory competence center

Allosteric modulation of the chaperone protein Hsp90
Hsp90 (heat shock protein 90) is a chaperone protein that assists the correct folding of other proteins, called clients. Hsp90 plays a pivotal role in the cell life cycle and it is crucial for the cell response to stressful conditions. As many Hsp90 clients are oncoproteins, this chaperone is nowadays an established target in cancer therapy. From a structural point of view, Hsp90 is a homodimer and each monomer is constituted by three domains, an N-terminal Domain (NTD), where the main ATP-binding site is located, a Middle Domain (MD) and a C-terminal Domain (CTD), where the dimerization takes place.
The majority of the small molecules developed targeting Hsp90 aim to displace the ATP from its binding site, blocking its ATPase and therefore chaperoning activity altogether. Unfortunately, this block triggers the Heat shock cell-survival mechanism, with increased expression of Heat shock proteins, including Hsp90 and Hsp70.
Our aim is instead to tune the activity of the chaperone with molecules targeting an allosteric site located between the MD and the CTD, close to the dimerization interface and recently discovered.
We synthesised a family of about 60 compounds based on the natural product Eupomatenoid-2 that are able to accelerate the ATPase activity of Hsp90 with interesting downstream effects.
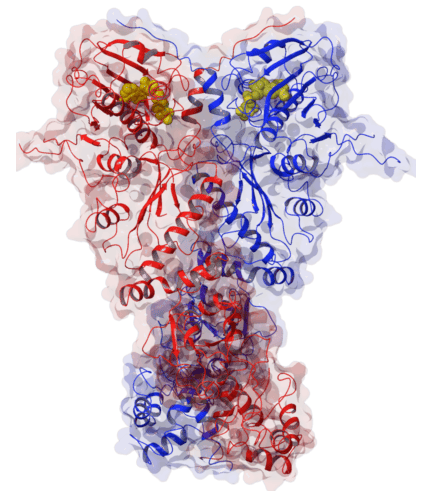
Figure 1: X-ray crystal strucuture of yeast Hsp90 in its closed conformation (2cg9.pdb) shown in complex with ATP (yellow spheres).
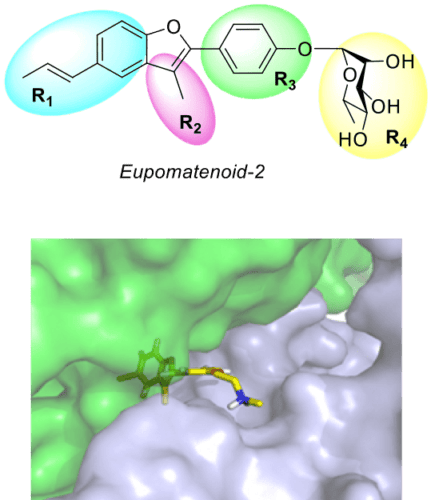
Figure 2: top) the natural compound Eupomatenoid-2 and the four diversification areas we tackled over the years. Bottom) One benzofuran derivatives that activates Hsp90 ATPase activity, docked within the allosteric site close to the C-terminal domain.
